Screening of yeasts obtained from different fermented foods for their ability to produce pectinase
Abstract
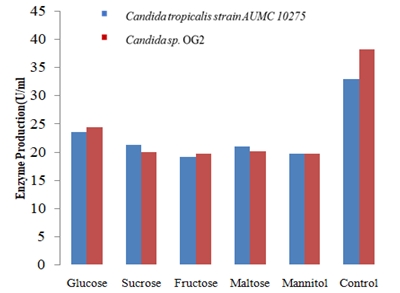
In the present study, Citrus pectin was used for the production of pectinase enzyme by yeast isolates using submerged fermentation. Fifty yeasts were isolated from different fermented foods and screened for their producing ability. Candida sp. OG2 and Candida tropicalis strain AUMC 10275 were the yeast isolates with the best potential of pectinase production. Fermentation parameters such as incubation period, pH, temperature, carbon and nitrogen source were optimized under submerged fermentation. The optimal conditions for pectinase production were found to be incubation time 48 hours, pH 6.0 and temperature 40°C. Citrus pectin best induced the production of pectinase while yeast extract/peptone (1:1) was the best source of nitrogen. Pectinase produced by Candida tropicalis strain AUMC 10275 was purified at 4.00 folds with a specific activity of 63.99 U/ml. The yeasts obtained from fermented foods have the ability to produce pectinase enzyme under optimized conditions and can be used for industrial purposes.
Downloads
References
2. Huch M, Franz CMAP. Coffee. In: Advances in fermented foods and beverages. Woodhead Publishing: Sawston, UK, 2015: 501-513.
3. Sakiyama NS, Ferrao MAG. Botany and production of coffee. In: Cocoa and coffee fermentations. Schwan RF, Fleet GH, eds. CRC Press: Boca Raton, FL, USA, 2015: 341-365.
4. Martos MA, Zubreski ER. Isolation of a yeast strain able to produce a polygalacturonase with maceration activity of cassava roots. Food Sci Technol. 2013; 33: 332-338.
5. Rattan S, Parande AK, NagarajuVD, Ghiwari GK. A comprehensive review on utilization of wastewater from coffee processing. Environ Sci Pollut Res. 2015; 22: 6461-6472.
6. Haile M, Kang WH. The role of microbes in coffee fermentation and their impact on coffee quality. J Food Quality. 2019: ID 4836709.
7. Romero Cortes T, Cuervo-Parra JA, Jose Robles-Olvera V, Rangel Cortes E, Lopez Perez PA. Experimental and kinetic production of ethanol using mucilage juice residues from cocoa processing. Int J Chem React Eng. 2018; 16(11): 20170262.
8. DeBruyn F, Zhang SJ, Pothakos V, Torres J, Lambot C, Moroni, AV, DeVuyst L. Exploring the impacts of postharvest processing on the microbiota and metabolite profiles during green coffee bean production. Appl Environ Microbiol. 2017; 83: 98.
9. Kurtzman CP, Fell JW. The yeasts - a taxonomic study, 4th edn. Elsevier Science, Amsterdam. The Netherlands. 1998.
10. Heritage J, Evans EGV, Killington R. Introductory Microbiology. 2000: 100-102.
11. Van der Walt JP, Yarrow D. Methods for the isolation, maintenance, classification and identification of yeasts. In: The yeasts - a taxonomic study. Kreger-Van Rij. Elsevier Science Publishers. 1984: 47-104.
12. Gadaga TH, Mutukumira AN, Nanhus JA. Enumeration and identification of yeasts isolated from Zimbabwean traditional fermented milk. Int Dairy J. 2000; 10: 459-466.
13. Arnold K, Marko L, Kersti K. Extraction of genomic DNA from yeasts for PCR-based application. Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia Bio Techniques. 2011; 50: 325-328.
14. Oskay M, Yalcin HT. Screening of yeast strains for pectinolytic activity: Effects of different carbon and nitrogen sources in submerged fermentations. Online J Biol Sci.2014; 15(3): 89-96.
15. Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959; 31: 426-428.
16. Khairnar Y, Krishna K, Boraste A, Gupta N. Study of pectinase production in submerged fermentation using different strains Aspergillus niger. Int J Microbiol Res. 2009; 1: 13-17.
17. Dixon M, Webb EG. Enzymes. 2nd edn. Academic Press. Inc. New York. 1964.
18. Moyo S, Gashe BA, Collison EK, Mpuchane S. Optimising growth conditions for the pectinolytic activity of Kluyveromyces wickerhamii by using response surface methodology. Int J Food Microbiol. 2003; 85: 87-100.
19. Hoa BT, Hung PV. Optimization of nutritional composition and fermentation conditions for cellulose and pectinase production by Aspergillus oryzae using response surface methodology. Int Food Res J. 2013; 20: 3269-3274.
20. Chellegati MC, Fonseca MJV, Said S. Purification and partial characterization of exo-polygalacturonase from Mucor rouxii NRRL 1894. Enzyme Microbiol Technol. 2002; 41: 800-805.
21. Freitas PMN, Martin D, Silva R, Gomes E. Production and partial characterization of polygalacturonase production by Thermophilic monascus sp. N8 and by thermo tolerant Aspergillus sp. N12 on solid state fermentation. Braz J Microbiol. 2006; 37: 302-306.
22. Helms D, Helms C, Kosinski R, Cummings J. Biology in the laboratory. 3rd edn. Wilson J, ed. Freeman Publishing, New York, New York.1998; 9: 1-8.
23. Roosdiona A, Sasangka P, Chanif M. Production and characterization of Bacillus firmus pectinase. J Pure Appl Chem Res. 2013; 2(1): 35-41.
24. Anosike EO. Basic enzymology. University of Port Harcourt Press. 2011: 11-87.